Copyright © 2007-2010 BiochemLabSolutions.com
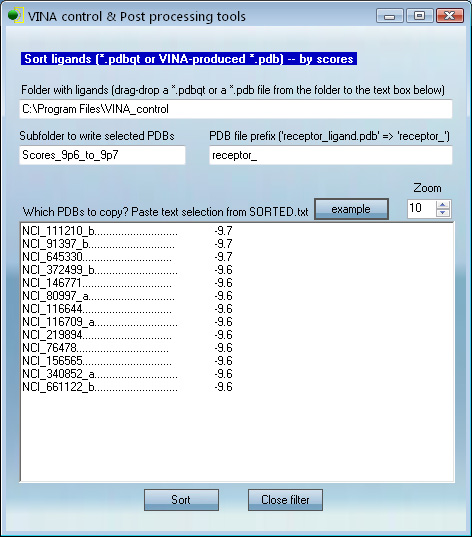
VcPpt contains tools for sorting any number of docked PDBs by energy. The input is a folder with Vina-produced PDB files, the output is a subfolder with PDB files that have docking score in a chosen range.

This utility helps find the best binders from in silico high throughput screening. An in silico screen may result in 10,000+ pdb files, most of which will have poor scores and need to be omitted from further analysis. Here one can find top binders and copy them to a new subfolder through a simple procedure.
1. Provide actual folder with screening results in place of "C:\Program File..." .
2. Set subfolder name for placing filtered PDB results in place of "Scores_9p6..."
3. PDB files from docking/screening run have file names in the form
"receptor_ligand1.pdb",
"receptor_ligand2.pdb",
"receptor_ligand3.pdb",
.....
"receptor" is the name of the protein PDBQT file used in the screening
"ligand1", "ligand2".... are the names of the small molecules PDBQT
At this step the program will need to know the protein name.
For example, If the screening was done with "kinase.pdbqt", "kinase_" should be typed instead of "receptor_".
4. Main text box contains the filtering rules. Ligands listed there provide the guide for
finding and copying PDBs to the filtered subfolder.
The whole-ligand list is automatically generated during the docking run and is called "SORTED.txt". One should open SORTED.txt in a text editor and copy any number of entries (for example, top 100) to the filtering rules window. When copying, avoid empty lines at the list's end. Each line should have a ligand.
5. The "Sort" button will run the sorting job.
If "SORTED.txt" is not available. For example, you combined separate VcPpt screening results into one folder, or have Vina docking results made not with VcPpt.
1. In VcPpt, click "Radius"
2. Drag-Drop one of the PDBs to the orange pad (this will point VcPpt to the right working
directory with Vina outputs)
3. In "Make SORTED.txt for any folder with VINA PDBs", PDBQT protein name with "_" goes
instead of "receptor_" (as in step 3 above)
4. "Make" will prepare new "SORTED.txt" in the docked ligands directory
If PDB docking files were made not with VcPpt, these PDB files must be renamed to contain a common prefix. For example if one's Vina output files at hand are: "drug.pdb", "adenine.pdb", "19000.pdb", one renames them to "new_drug.pdb", "new_adenine.pdb", "new_19000.pdb" and replaces the word "receptor_" with "new_" in the text box of "Make SORTED.txt for any folder with VINA PDBs".
The "Make" button will produce "SORTED.txt".
_____________________
All sorting is done based on the top docked solution in the PDB files. By combining the sorting by scores and sorting by binding position it takes minutes to find the best binders to any given pocket among thousands of docking results.
1. Provide actual folder with screening results in place of "C:\Program File..." .
2. Set subfolder name for placing filtered PDB results in place of "Scores_9p6..."
3. PDB files from docking/screening run have file names in the form
"receptor_ligand1.pdb",
"receptor_ligand2.pdb",
"receptor_ligand3.pdb",
.....
"receptor" is the name of the protein PDBQT file used in the screening
"ligand1", "ligand2".... are the names of the small molecules PDBQT
At this step the program will need to know the protein name.
For example, If the screening was done with "kinase.pdbqt", "kinase_" should be typed instead of "receptor_".
4. Main text box contains the filtering rules. Ligands listed there provide the guide for
finding and copying PDBs to the filtered subfolder.
The whole-ligand list is automatically generated during the docking run and is called "SORTED.txt". One should open SORTED.txt in a text editor and copy any number of entries (for example, top 100) to the filtering rules window. When copying, avoid empty lines at the list's end. Each line should have a ligand.
5. The "Sort" button will run the sorting job.
If "SORTED.txt" is not available. For example, you combined separate VcPpt screening results into one folder, or have Vina docking results made not with VcPpt.
1. In VcPpt, click "Radius"
2. Drag-Drop one of the PDBs to the orange pad (this will point VcPpt to the right working
directory with Vina outputs)
3. In "Make SORTED.txt for any folder with VINA PDBs", PDBQT protein name with "_" goes
instead of "receptor_" (as in step 3 above)
4. "Make" will prepare new "SORTED.txt" in the docked ligands directory
If PDB docking files were made not with VcPpt, these PDB files must be renamed to contain a common prefix. For example if one's Vina output files at hand are: "drug.pdb", "adenine.pdb", "19000.pdb", one renames them to "new_drug.pdb", "new_adenine.pdb", "new_19000.pdb" and replaces the word "receptor_" with "new_" in the text box of "Make SORTED.txt for any folder with VINA PDBs".
The "Make" button will produce "SORTED.txt".
_____________________
All sorting is done based on the top docked solution in the PDB files. By combining the sorting by scores and sorting by binding position it takes minutes to find the best binders to any given pocket among thousands of docking results.

Using docking scores to select best binding ligands